Codon Optimization
Codon Optimization to Maximize Protein Expression
At ATUM, we strive to express your active protein at the highest levels. This requires selecting the best coding sequence for a gene. There are a vast number of possible coding sequences for a given protein – an average protein can be coded for in more ways than there are particles in the universe!
Codon usage within a gene is critical in determining the achievable protein expression levels. Certain sequences can be translated more readily by certain hosts, so selecting the best possible codons in the best possible order for a given host is crucial to maximising expression. Optimizing protein expression therefore requires choosing from an enormous number of possible DNA sequences. Our empirically-derived codon optimization GeneGPS technology uses state-of-the-art machine learning to select the best possible combinations of codons, ensuring that your gene is designed for efficient translation in your chosen host.
Codon Optimization Powered by GeneGPS®
Not all codon optimization tools are created equal. Most available codon optimization software are guided by theory or mimicry of natural gene characteristics predicted to increase expression. While they can offer increased expression with some targets, success is variable and does not offer prediction of which gene sequence will be the highly-expressed.
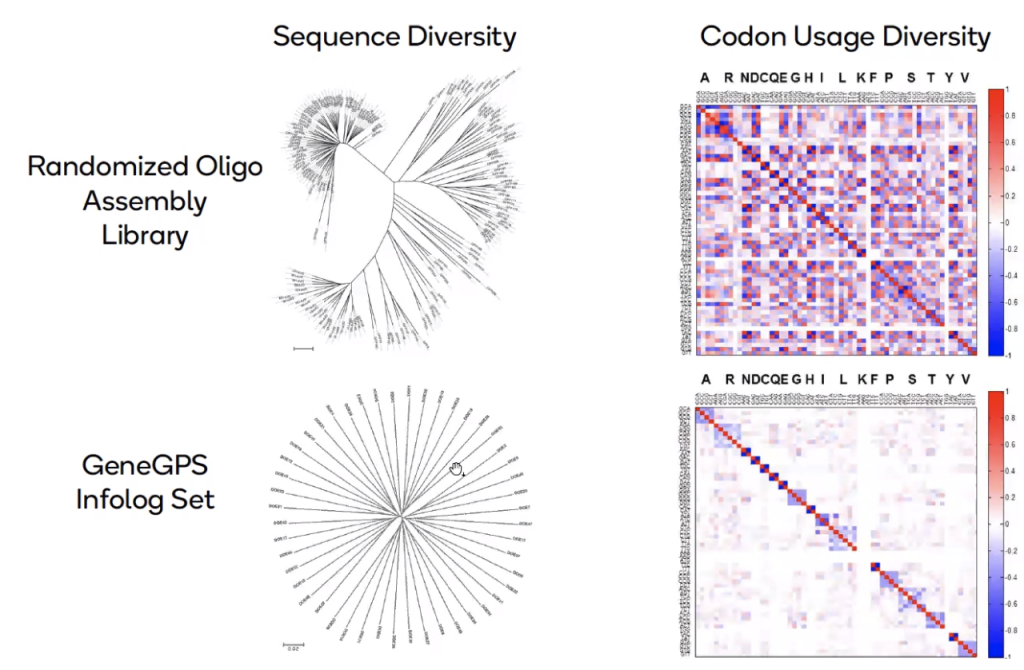
Our proprietary GeneGPS technology uses models based on our empirical learning about what works best in real world expression systems, and provides more accurate codon usage analysis predictions of the optimal sequence than models based on theory alone. Genes optimized with GeneGPS algorithms for maximal expression yield between 10 and 100-fold more protein while using smaller culture volumes and saving you months of time on painstaking optimization studies.
The left-hand panel shows protein expression data compared to predictions from traditional optimization thinking of using ‘most common codons’ (Codon Adaptation Index, CAI), which gives a poor correlation to yield. The right-hand panel instead shows ATUM’s approach. Data is modelled using machine learning methods to analyse the impact of different codon biases and other factors on the variation present in the data. This approach allowed us to build a model that made sense of the expression data and generate a more accurate and predictable view of codon usage.
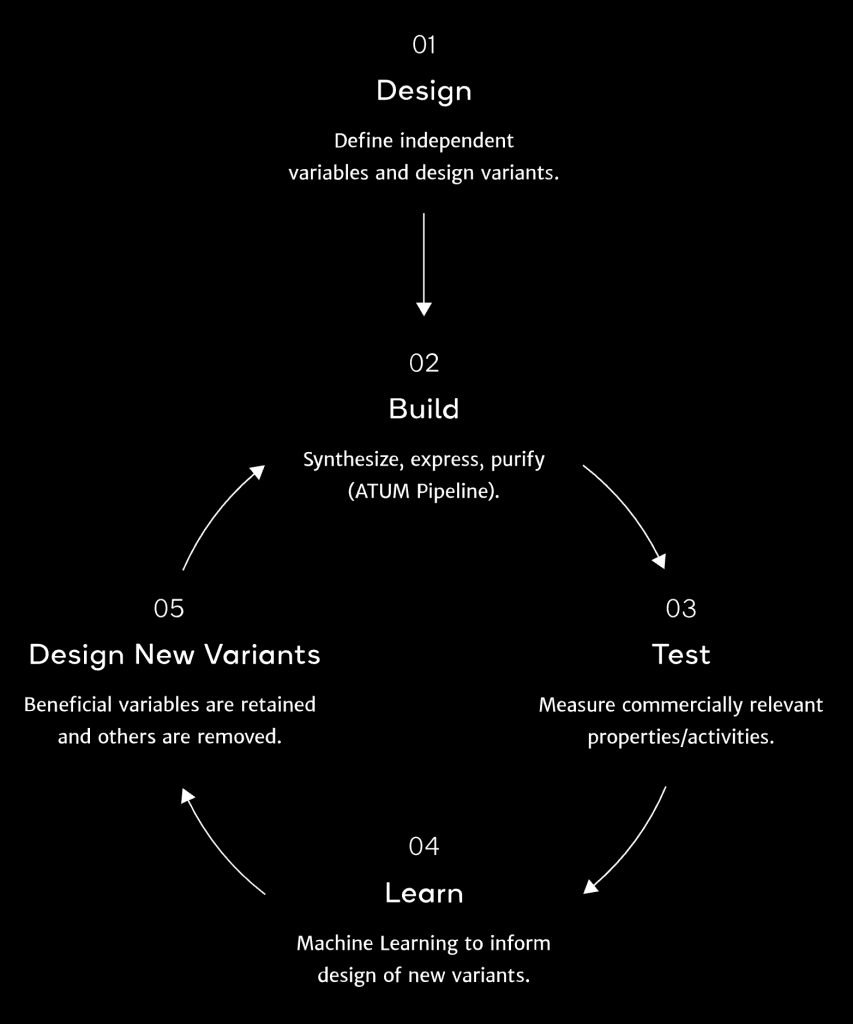
How GeneGPS Works
Codon optimization is the first step of ATUM’s Cell Line Development Services. We use our codon optimization algorithms, secretion signal toolbox and customized vector configurations to generate high productivity stable CHO-K1 cell lines. Expression of proteins in mammalian cells may be limited by codon usage. This is why our process begins with mammalian codon usage analysis and optimization to ensure we have the optimal sequence for maximal expression in the host.
Interrogating host preferences using multivariate machine learning and sequence space exploration is used to develop the GeneGPS algorithms, providing significantly increased expression over both wild type and traditional codon optimization methods. Multiple rounds provide magnitudes of functional improvement. See publications in the Resources for detailed scientific explanations.
Resources
- Design parameters to control synthetic gene expression in Escherichia coli Experimental determination of design parameters that formed the foundation of GeneGPS.
- Engineering genes for predictable protein expression Review of the GeneGPS gene optimization strategy and vector design.
- Developing codon optimization design for vaccine in Nicotiana benthamiana Webinar with Medicago - Codon optimization design for vaccines.
- Engineering genes for predictable protein expression