Protein from Mammalian Cells
Our experience in offering protein expression and cell line development services has translated into extensive expertise in vector design, and in understanding how vectors interact with their hosts to affect the quantity and quality of expressed protein. Below are examples of the approaches we apply as part of our customized protein services to optimize the expression of your protein from mammalian cells.
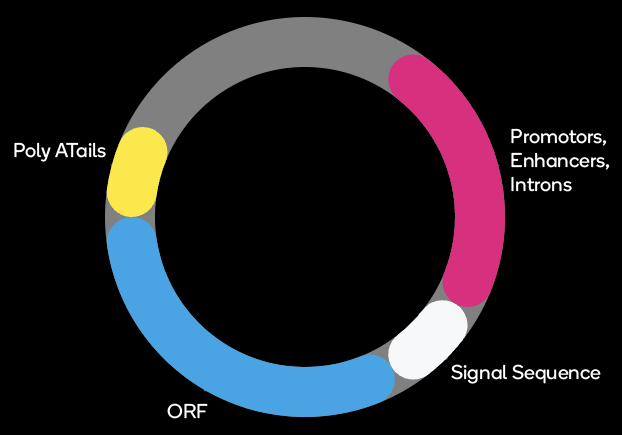
Common Vector Elements
Recording Open Reading Frame
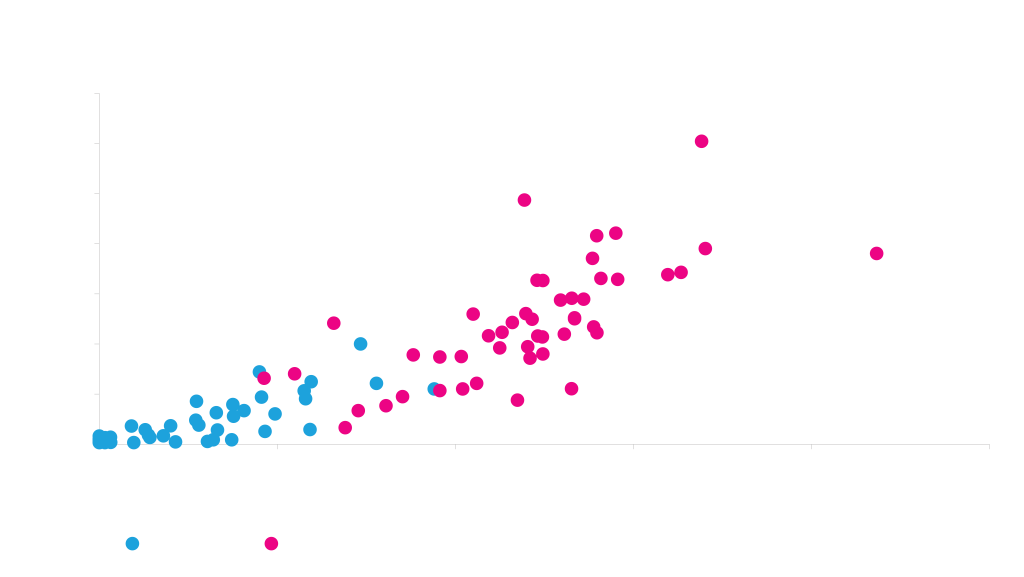
We have designed and expressed tens of thousands of genes in mammalian cells. By feeding this data into our GeneGPS platform, have identified critical sequence parameters that most strongly affect expression. The graph below shows a strong correlation between the expression of genes predicted by our design algorithm, and the expression level actually observed in HEK cells. This mammalian re-coding enables us to computationally design genes that will express well in the real world.
Signal Sequence
Mammalian cells are frequently used to produce secreted proteins. However, the effectiveness of a mammalian secretion signal sequence is highly dependent on the protein being secreted: signal sequences that work well for one chain of an antibody may be mediocre for another. We have identified many signal sequences and characterized their effectiveness across a range of proteins, including antibody heavy and light chains of different origins.
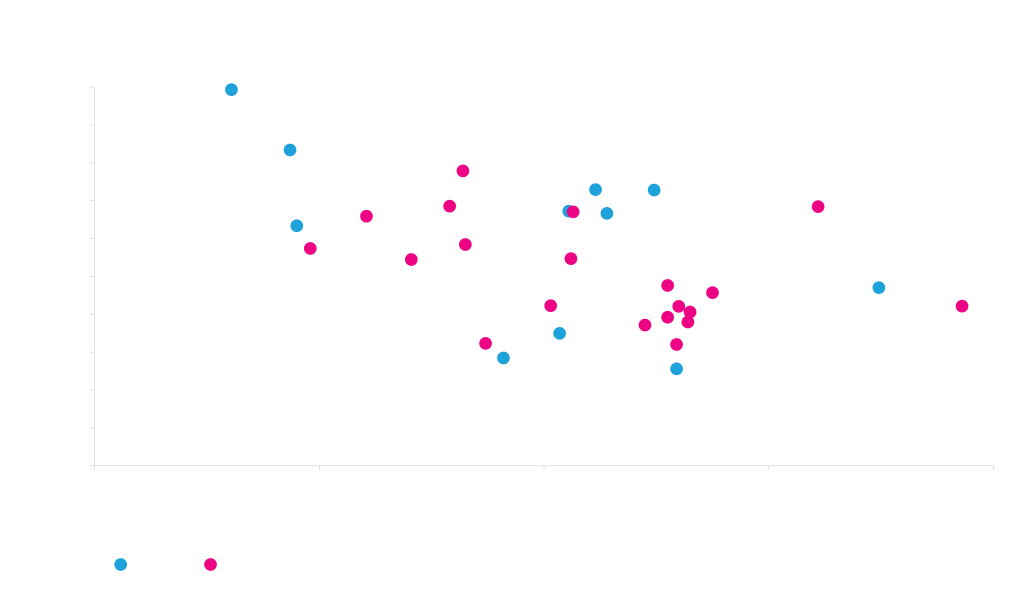
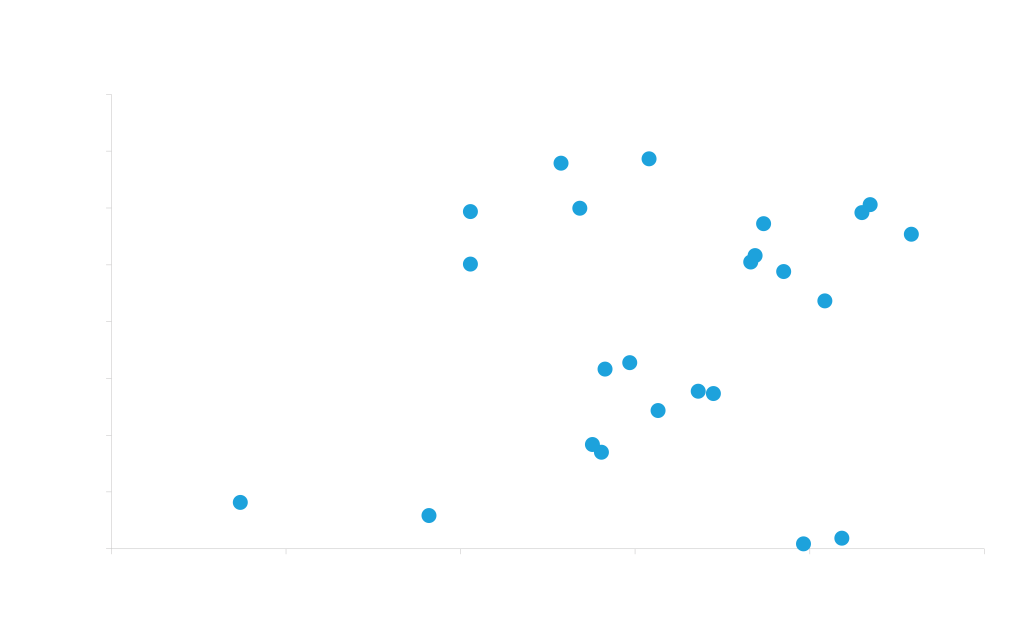
The antibody-dependence of secretion signal performance is exemplified in the graph below. By using different secretion signals for the heavy and light chains of two different antibodies, we observed a five-fold range in expression levels for Rituxan, and a more than twenty-fold range for Trastuzumab. There is also relatively little correlation between the secretion signals that work well for the two different antibodies. We therefore performed a more extensive study to identify generally good secretion signals.
Effectiveness of secretion signal is highly dependent on protein being secreted
Two different antibodies (Rituxan, Trastuzumab) were transiently expressed in HEK cells using 24 different heavy and light chain signal sequence combinations. Each data point represents the same vector and same signal sequence combination used for both chains of both antibodies.
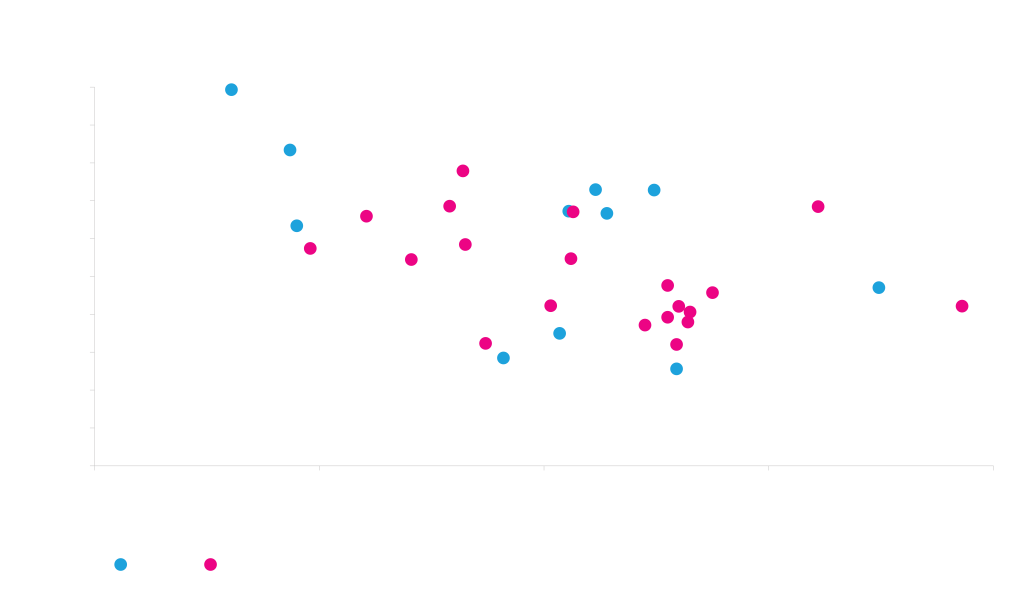
Effective secretion signals identified by machine learning algorithms.
Light Chain
Heavy Chain
Five different heavy and light chains were each fused to 23 different signal sequences and were expressed in various combinations from otherwise identical vectors in HEK293 cells. Machine learning was used to analyze the data and assess the effectiveness of each secretion signal. The more positive the regression weights, the more effective the secretion signal. This allowed us to identify secretion signals that are generally robust.
Vector Elements
Promoters, Enhancers, Introns
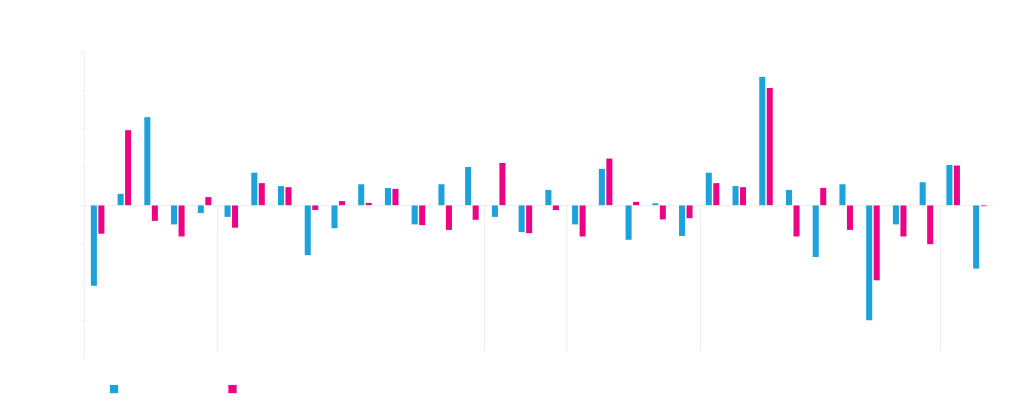
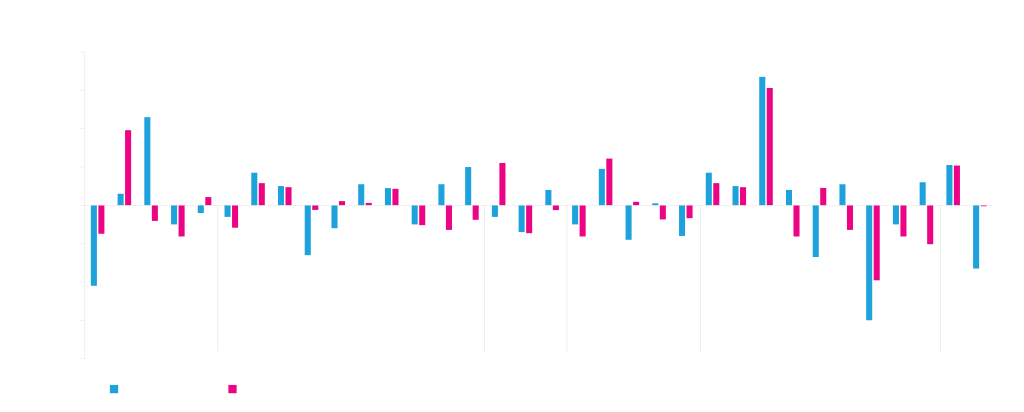
ATUM uses a variety of viral and cellular promoters as part of our protein expression services. These are combined with introns, which often contain enhancer sequences. Intron processing also enhances mRNA export from the nucleus into the cytoplasm for translation. Many of these elements have been modified from natural sequences during mammalian codon optimization to improve their function.
Twenty-four different vector elements were used in different combinations to build 40 different vectors. Transient expression of a test protein was then measured in HEK293 and ExpiCHO cells. The expression data was analyzed using machine learning. Elements that are beneficial for expression have positive regression weights, negative weights indicate a deleterious effect on expression. The magnitude of the weight corresponds to the magnitude of the effect of the element. Although the vector elements generally perform comparably in ExpiCHO and HEK293, there are sometimes differences (see introns and viral replication elements).
PolyA Tails
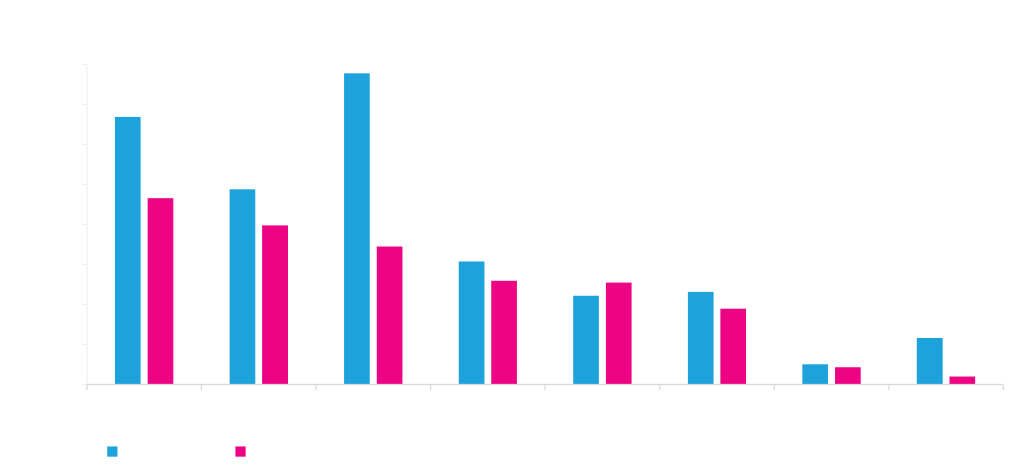
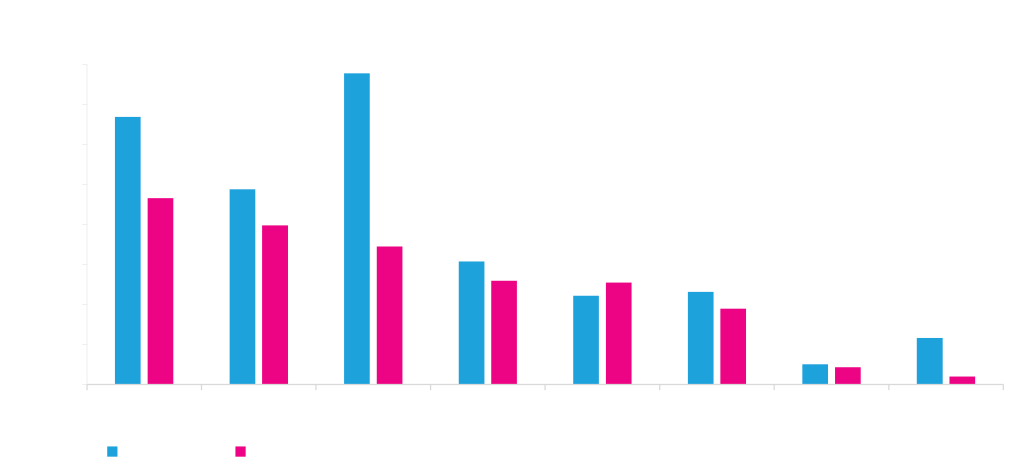
Polyadenylation serves to protect the mRNA from degradation. PolyA tails also assist in the export of mRNA from the nucleus to the cytoplasm. Polyadenylation therefore improves translation and the total amount of expressed functional protein. We have found that the choice of polyA signal sequence has a profound effect on mammalian protein expression.
Choice of polyA signal affects mammalian protein expression
The graph shows expression of GFP driven by the same promoter and terminated by eight different polyadenylation signal sequences. Although the polyA signals generally perform comparably in HEK and ExpiCHO, there are sometimes differences (A and C above).
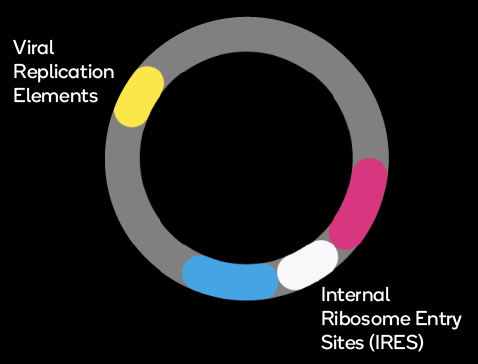
Viral Replication Elements
Following transfection, plasmids that are not integrated into a host chromosome, are lost by degradation and by dilution as cells divide. Viral replication elements can slow this loss, thereby enhancing protein expression. Transient HEK and CHO cells have different preferences for these elements.
Internal Ribosome Entry Sites (IRES)
IRES (Internal Ribosome Entry Site) elements create a second, cap-independent site for ribosome binding, resulting in independent, bicistronic expression of two ORFs from a single mRNA molecule. The ratio at which the two proteins are expressed depends on the efficiency of ribosome entry at the IRES relative to ribosome binding 5′ end of the mRNA. As part of our protein expression services or VectorGPS services, we have developed a set of novel IRES elements; each produces a defined expression ratio between the first and second protein in every transfected cell.
Choice of IRES elements to control the ratio of bicistronic expression between two proteins
Different IRES elements were incorporated into a GFP-IRES-RFP configuration, in otherwise identical expression vectors. Expression of GFP and RFP in HEK293 and ExpiCHO cells was measured 72 hours post-transfection. IRES efficiency is the ratio of RFP:GFP.


Stable Expression of Multiple ORFs
Controlling multi-ORF expression
We use our proprietary Leap-In technology for the construction of stable cell lines. Leap-In transposase can integrate expression constructs harboring up to four transcriptional units into a host genome with no rearrangements. By selecting the appropriate vector elements, we can control the relative expression ratios of up to four protein subunits within the same cell.
Leap-In Transposon 2 ORFs
Leap-In Transposon 3 ORFs
Leap-In Transposon 4 ORFs
Selection Markers
A variety of selectable markers are available for conferring resistance to puromycin, neomycin, hygromycin and blasticidin. Alternatively, genes encoding glutamine synthetase and dihydrofolate reductase can be used for drug-free selection.
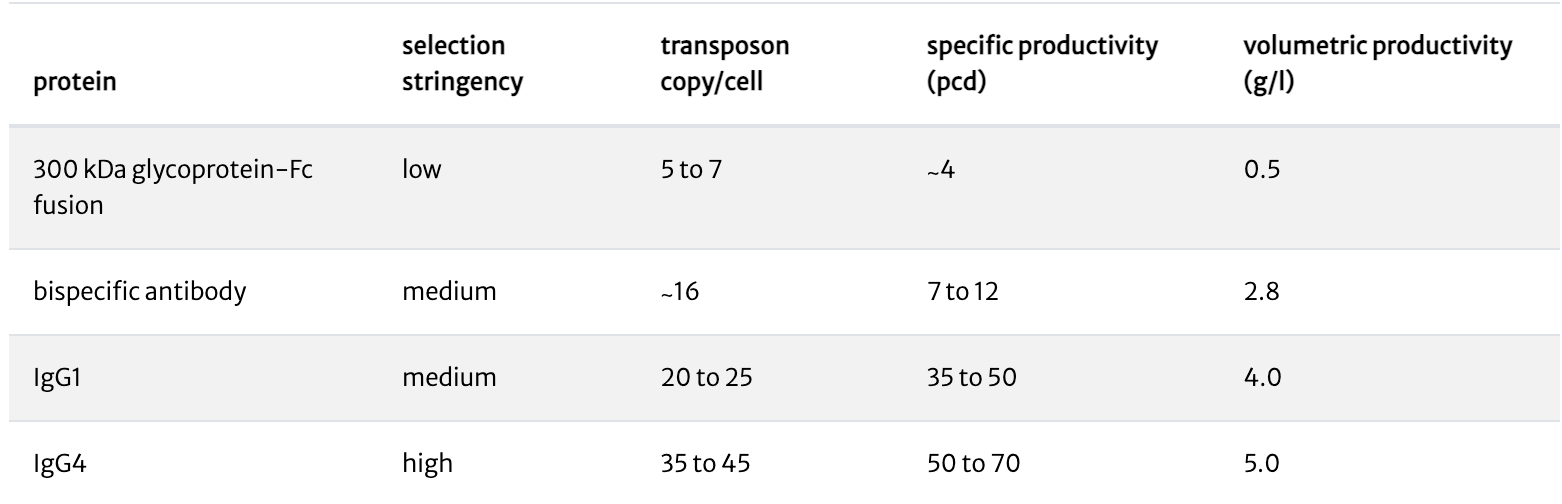
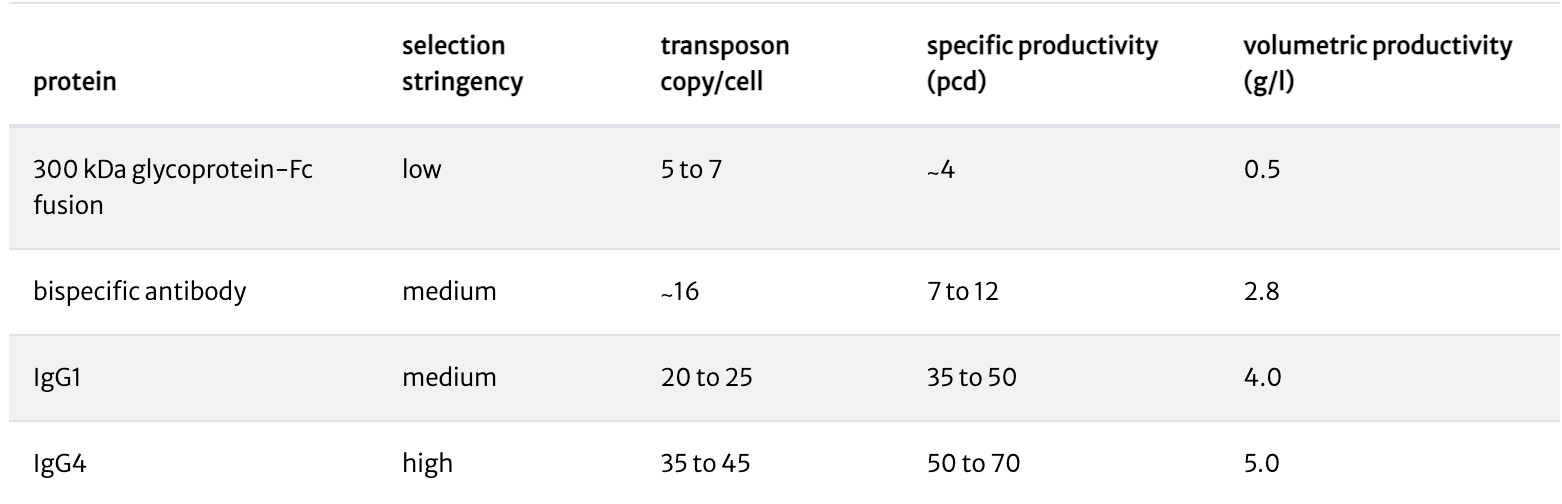
We have developed several attenuated selectable markers through which selection stringency can be manipulated. Under higher selection stringency, only cells with more copies of the selectable marker, and thus of the expression construct, will survive. For low toxicity molecules (such as monoclonal antibodies), high selection stringency results in highly productive stable cell lines and pools. For challenging or toxic molecules, choosing the appropriate selection stringency enables the creation of stable cell lines optimized for the expression of that molecule.
Under high selection stringency, stable pools express ≥5 g/l of human monoclonal antibodies. Lower selection stringencies result in stable pools with lower productivities.
Under medium selection stringency, stable pools express 2.8 g/l of a bispecific antibody. Cells did not survive more stringent selection conditions.
Under low selection stringency, stable pools express ∼0.5 g/l of a 300kDa glycoprotein-Fc fusion. No cells survived medium or highly stringent selection conditions.
Protein Expression Optimization
In addition to vector elements and cell lines described, ATUM further optimizes protein expression using a range of conditions including temperature and special media additives.
Transient Vector Performance Varies with Cell Type
DasherGFP Expression in Transiently Transfected HEK293 and CHO Cell Lines
Cell Lines
Individual vector elements, and therefore entire vectors, can behave differently across even closely related cell lines. By understanding the ‘preferences’ of each cell line, we can design the vector most appropriate for that cell line. We have already build vectors optimized for HEK293 and CHO expression hosts, used both in-house at ATUM and also by clients in their proprietary cell lines. In addition to catering for clients working with CHO cell lines, we routinely create new vectors for clients working in non-CHO cell lines with applications in cell therapy such as the Jurkat/T-cell line.
DasherGFP expression in transiently transfected cells. HEK293 (adherent), Expi293® (suspension), CHO-K1 (adherent), Freestyle® CHO-S (suspension), and ExpiCHO® (suspension) cells were transfected at 2 x 106 cells/ml using Lipofectamine 2000; transfections were carried out in triplicate and incubated for 72 hours post-transfection. Cells were lysed using M-PER and expression measured on a fluorimeter. Orange and grey data points are shown for comparison.