Protein Engineering
ProteinGPS® is an engineering technology that optimizes proteins for real world applications. A small number of variants are tested using high quality assays representative of the functionality, developability and manufacturability properties that your protein will need. Iterative machine learning and testing steps result in an engineered protein that will work where it matters: the real world.
REAL WORLD PERFORMANCE
High-quality low-throughput assays more precisely mimic the conditions that your protein will experience during its lifecycle. The result is a product that performs better in the real world, from manufacturing to its end-application.
SPEED TO MARKET
ATUM’s efficient workflow integrates in-house-media, assay, analytical and process development expertise. This accelerates project timelines and positions your protein for fast scale-up and manufacturing.
INTELLECTUAL PROPERTY
We can help you understand the IP landscape surrounding your protein. This translates to a stronger intellectual property position, giving you freedom to operate and broader patent claims, thereby reducing the risk of product obsolescence and being quickly overtaken by your competitors.
Bioprospecting for Better Starting Proteins
You want to engineer a protein, but you are not sure about the best starting point? We use phylogenetic analysis to identify, build, and test many orthologs of your protein to find variants with better starting properties for protein engineering. Every time we do this, we identify a preferable starting protein and generate additional data that can be used to find amino acid substitutions encoding functional properties and data to strengthen your intellectual property portfolio.
-
ProteinGPS® Variant Design
Permits Real World Testing
Natural selection guides our ProteinGPS choice of amino acid substitutions.
We begin with a sequence alignment of natural homologs to your protein. Then we score and rank every possible amino acid substitution using several different evolutionary, structural and functional parameters. Finally, we apply Design of Experiment (DoE) to design protein variants incorporating the highest ranked substitutions. Fewer than 100 variants are needed for each ProteinGPS iteration.
Traditional library screening strategies are time consuming, expensive and inefficient. With millions of variants, you are restricted to using high-throughput screens that typically do not accurately measure protein properties under real-world conditions. With fewer than 100 variants, ProteinGPS allows us to use low-throughput, high-quality assays that mimic the real-world conditions under which you need your protein to perform.
- Machine Learning Tells Us Which Sequence Changes Affect Protein Properties
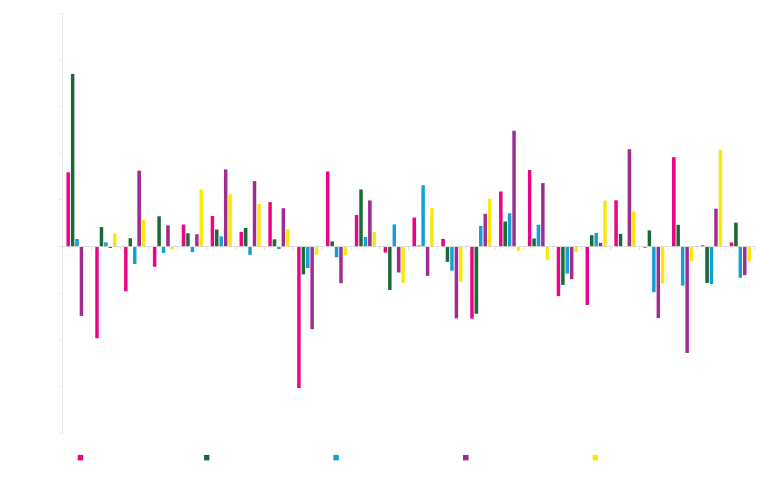
Once the functional properties that are important for the intended application have been measured, ProteinGPS® machine learning is used to determine the causal effect of each sequence change. The following graph shows the impact of 48 different changes on 3 properties of an industrial enzyme. It is striking that very few changes have a positive effect on activity, thermostability and solubility simultaneously. This is a severe limitation in traditional library screening approaches.
When one property is measured first, and all library members not improved in that dimension are discarded, improvements in other important properties are lost. In contrast, ProteinGPS allows us to identify and combine changes that separately improve solubility, activity, and thermostability, resulting in an enzyme that is improved for all three.
Pfizer required improved stereospecific activity in an enzyme used for the synthesis of a pharmaceutical intermediary. After 4 rounds of engineering using fewer than 300 variants, the project goals were achieved with over 100-fold improvement in activity.