ProteinPaintbox
YIELD
Monitor inducible protein expression with ATUM’s fluorescent and chromogenic proteins.
SPEED
ATUM’s fluorescent and chromogenic proteins are an ideal source of protein coding sequences that can be quickly cloned into any expression vector of choice.
PRECISION
ATUM’s fluorescent and chromogenic proteins cloned into expression vectors can be used as positive controls.
ProteinPaintbox®
ProteinPaintbox®
Synthetic non-Aequorea fluorescent and chromogenic proteins bring a world of color to your research.
Overview
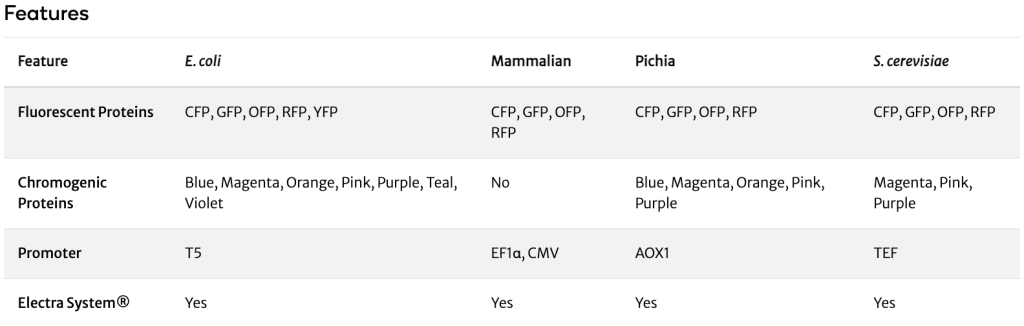
ATUM’s synthetic fluorescent and chromogenic proteins (non-Aequorea) are an ideal source of protein coding sequences (genes) that can be easily excised using the flanking restriction sites and cloned into any other expression vector of choice. These vectors can also be used as expression vectors or as positive controls, and allow monitoring of inducible protein expression.
Presentations, Posters and Whitepapers
- SB6.0 Conference Poster: ProteinPaintbox® – An Expansive Palette
- Dual or Co-expression data for ProteinPaintbox
- PrancerPurple Application: Bio-Rad untagged protein purification
- The Tech participates in iGEM – DNA2.0/ATUM sponsored
- U.S. Patent Nos. 8,975,042; 9,290,552; 9,493,521 and 9,771,402. Fluorescent and colored proteins and methods for using them. Minshull and Theodorou
Fluorescent Proteins - Overview
ATUM’s synthetic non-Aequorea fluorescent proteins are intended to be used as a source of different fluorescent protein coding sequences (genes) that can be easily excised using the flanking BsaI restriction sites and cloned into any other expression vector of choice. These vectors can also be used as expression vectors (T5 promoter) or as positive controls and allow monitoring of inducible protein expression.

Chromogenic Proteins - Overview
ATUM’s chromogenic proteins are intended to be used as a source of different color protein coding sequences (genes) that can be easily excised using the flanking BsaI restriction sites and cloned into any other expression vector of choice. These vectors can also be used as expression vectors (T5 promoter) or as positive controls and allow monitoring of inducible protein expression.

What are the terms of use?
See the complete legal Terms and Conditions.
Can you explain the protein names?
Yes! The individual color names in the ProteinPaintbox derive from our 2011 Holiday publication, Nine improved monomeric fluorescent proteins from Rangifer tarandus, where the proteins were named after Santa’s reindeer (Rudolph, Donner, Comet, etc…). As the ATUM fluorescent and chromogenic proteins became available for sale, we kept the reindeer names. As we added new colors to our product line, we continued with the holiday naming theme, branching out to include a diversity of cultural winter traditions, classic holiday literature, drama, and of course, television cartoons. We hope you enjoy this naming scheme. Should you have any questions regarding the protein names or wish to suggest additional names, please email us at communications@atum.bio.
Can you tell me more about the T5 promoter?
Sure, you can learn more about vector design with T5 promoter, lac operator for IPTG induction and increased protein expression in bacterial systems here.
What volume is provided?
2µg lyophilized plasmid
Why are there so many?
Multiple CFPs, GFPs, etc. have been selected to cover a range of intensities. For example, FrostyCFP – 20000, CindyLouCFP – 35000, TwinkleCFP – 12000. It is also important to remember, that while optimized for each host system, these protein sequences have not been optimized for your individual project needs. By providing you with a large range of choices, you can test a variety of proteins to determine which works best for your research.
Can Paintbox Proteins express in anaerobic systems?
No, the Paintbox proteins need oxygen to mature, so cannot be expressed anaerobically.
Why are the Biobrick FPs not included in your Paintbox?
The Biobrick set (3 FPs, EiraCFP, JuniperGFP and BlazeYFP) have been donated by ATUM to the BioBricks Foundation and are covered under the Biobricks licensing agreement (BLA). These belong to BioBricks, and are therefore not included in the ATUM ProteinPaintbox set.
Are these unique proteins? Or these essentially derivatives or mutants of a 'base' protein?
The green/yellow FPs share only 27% homology at the amino acid level. Thus, it is fair to say that they are all unique proteins.
How long is the maturation time?
In the ATUM labs, we get colored colonies in 24hrs. As for how fast the maturation is, we have not done a time-course analysis. So, we cannot say if color develops in an hour or if it takes longer. Maturation time at 37°C – 24 hrs incubation is fast, 24-72 hrs is medium.
Do you have a simple method to measure how many GFP molecules are expressed?
ATUM offers an antibody (anti-CometGFP, catalog #AB-01) that will detect GFP which could be used to determine the number of GFP molecules. However, we do not currently have data for this. Note, since this is a polyclonal Ab, it should be able to detect any of our GFPs.
Why can I detect one protein but not another?
Maturation time and intensity varies for each of the proteins.
For example, the intensity of RudolphRFP is lower than DasherGFP, while maturation time for RudolphRFP is longer than for DasherGFP. Thus at an early timepoint, you may have the appearance of excellent DasherGFP expression, while only minimal RudolphRFP expression.
What is the half-life of these proteins?
We have not measured half-life.
However, we have looked at color after a week and it appears to remain stable.
Do the proteins require any particular genetic background to mature?
These proteins should be able to express in any genetic background that will support T5 driven expression.
Can the proteins mature in minimal media?
Yes, they should mature in minimal media induced with IPTG.