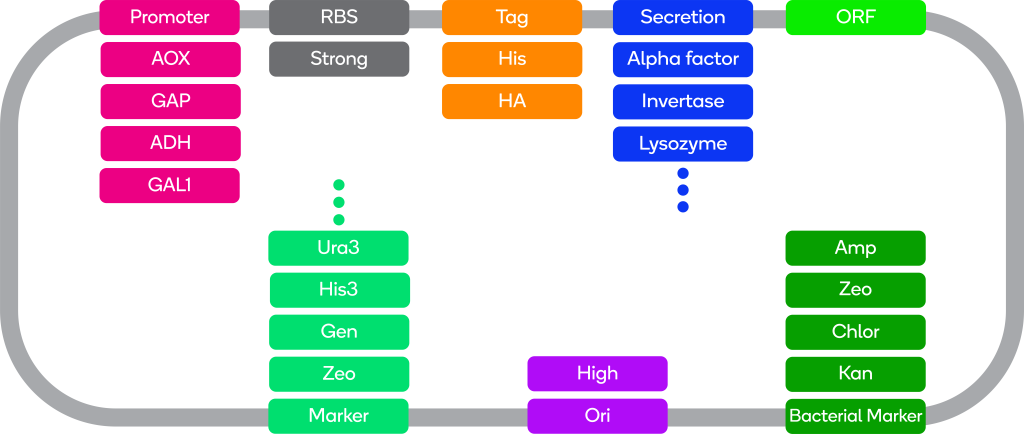
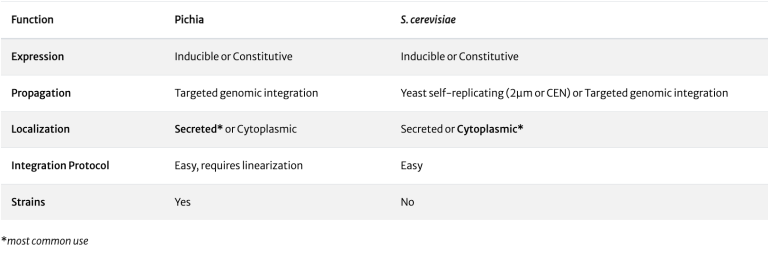
ATUM offers vectors for expression in the industrial production yeast Pichia pastoris (also called Komagataella phaffi), and for the well-understood model organism Saccharomyces cerevisiae.
YIELD
The methylotrophic yeast Pichia pastoris is a useful system for expression of milligram-to-gram quantities of protein for both basic research and industrial manufacturing.
SPEED
Test your gene in parallel in several ready-to-use vectors with a variety of properties most likely to increase expression.
PRECISION
Want the very best vector for your system? ATUM will create unique combinations and configurations of vector components using our machine learning technology, to create a custom vector exactly suited to your needs.
Pichia Pastoris Overview
The methylotrophic yeast Pichia pastoris is a useful system for the expression of milligram-to-gram quantities of protein for both basic laboratory research and industrial manufacturing. ATUM offers Pichia vectors with a choice of promoters for cytoplasmic or secreted expression.
The major advantage of expressing secreted heterologous protein is that Pichia secretes very low levels of native proteins. This means that the secreted heterologous proteins comprise the vast majority of the total protein in the media, which facilitates downstream protein purification. Secretion requires the presence of a secretion signal sequence to target the expressed protein to the secretory pathway. While several different secretion signal sequences have been used including the native secretion signals present on some heterologous proteins, success is dependent on the recombinant protein being expressed.
Advantages of Pichia for heterologous protein expression:
- Does not require complex media or culture conditions
- Genetically easy to manipulate
- Eukaryotic protein synthesis pathway
- Can be grown to very high cell densities in minimal media
- Integrated vectors help genetic stability of the recombinant genes
- High protein expression levels at the intracellular or extracellular levels
- Ability to perform higher eukaryotic protein modifications, such as glycosylation, disulphide bond formation and proteolytic processing
Available as Electra vectors for rapid cloning and testing protein expression in multiple vectors
Expression data
Effectiveness of Secretion Signals is Protein Dependent
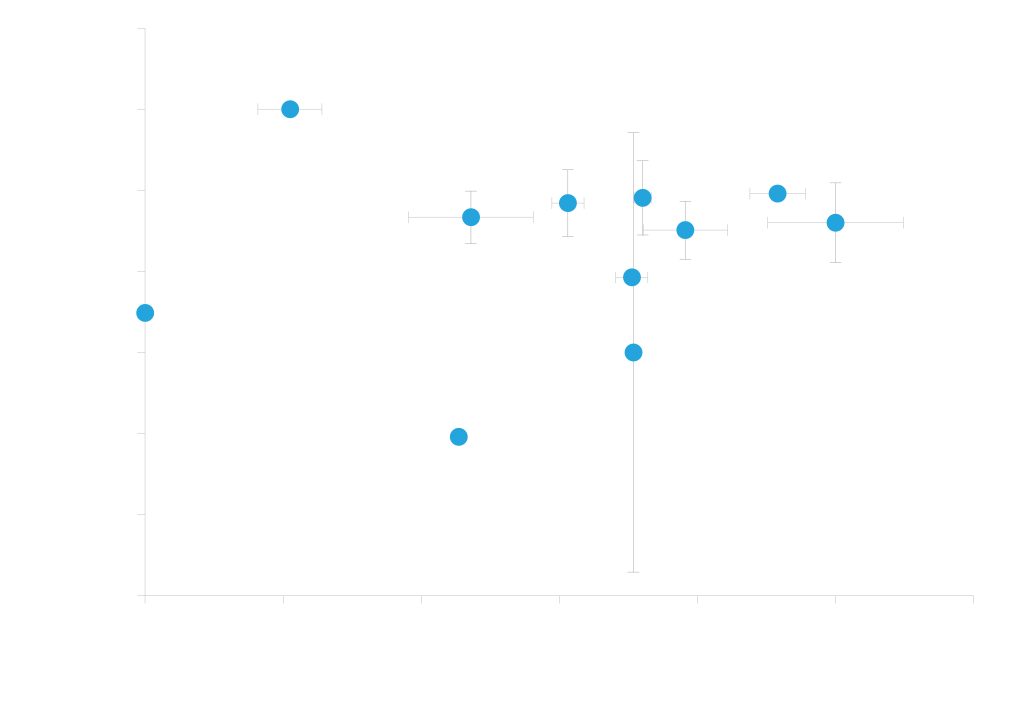
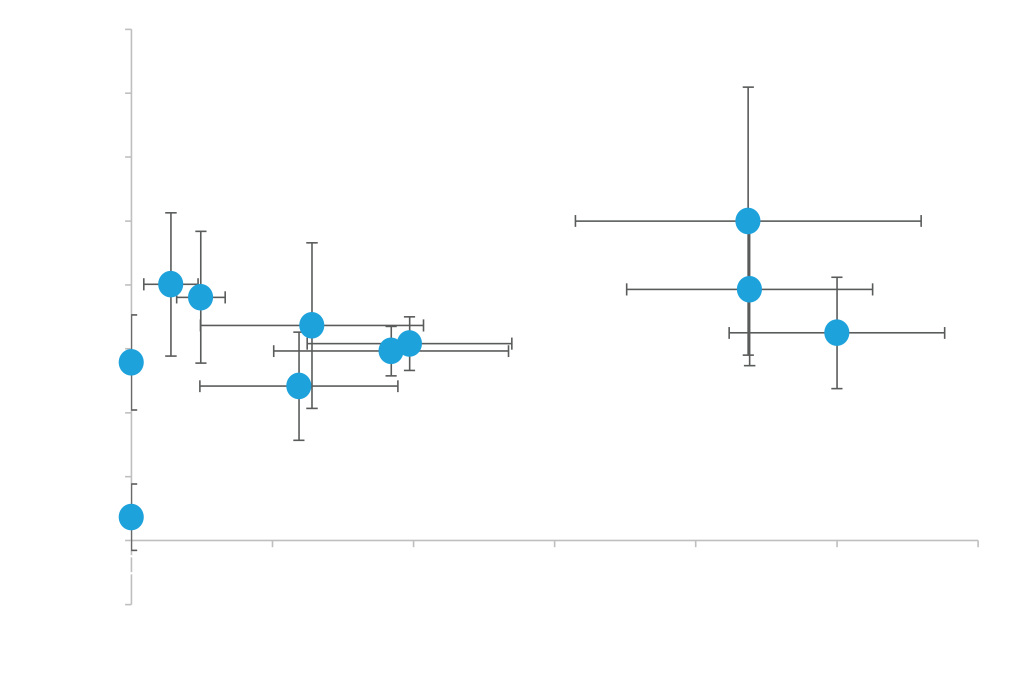
Efficient protein secretion is highly dependent on the combination of the protein and secretion signal used. We therefore offer vectors with eleven different secretion signals for targeting proteins to the secretory pathway. You can select vectors from the interactive graph below; we recommend testing the entire vector panel to identify the signal that works best with your protein.
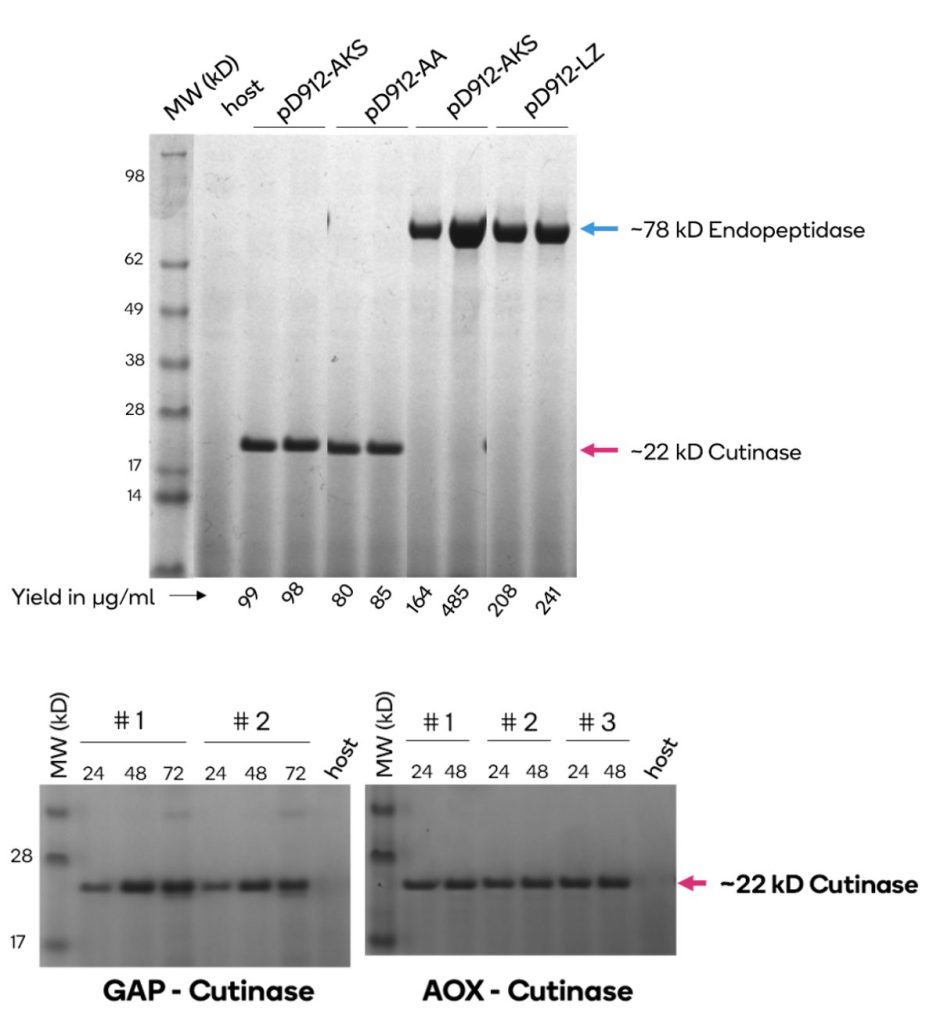
Different secretion signals work well with different proteins. Secreted expression of cutinase and endopeptidase driven by a methanol inducible AOX1 promoter and various secretion signals (pD912-XX). Expression values shown on x- and y-axis are measurements of expressed protein band densities from a SDS-PAGE gel using BSA as standard. In this example, the highest cutinase expression is observed with either alpha-factor secretion signal (AKS) or alpha-Amylase (AA) while the highest endopeptidase expression is observed with Lysozyme (LZ) secretion signal. Expression levels range from 20 to 100 µg/ml for Cutinase and 80 to 450 µg/ml for Endopeptidase; grown in small batch cultures.
Picha References
View Publications using ATUM Pichia Expression Vectors and/or Pichia host systems
Search the ATUM Literature Database, containing over 4,000 scientific publications using ATUM technology for references relevant to your research.
Learn more from these presentations and references:
- Pichia 2012 Conference Poster: High level protein expression in Pichia pastoris combining synthetic promoters and synthetic genes. Mellitzer, et al.
- Applied Synthetic Biology Europe 2012 Presentation by Anton Glieder: Synthetic promoters for recombinant protein expression in yeasts.
- Metab Eng 2010. Engineering the Pichia pastoris methanol oxidation pathway for improved NADH regeneration during whole-cell biotransformation. Schroer et al.
- Microb Cell Fact 2010. Stepwise engineering of a Pichia pastoris D-amino acid oxidase whole cell catalyst. Abad et al.
- Systems and Synth Biol 2010. Variable production windows for porcine trypsinogen employing synthetic inducible promoter variants in Pichia pastoris. Ruth et al.
Pichia Protocols
Competent Pichia yeast cell preparation protocol can be found here.
Pichia Culture and Induction Protocol from ATUM.
Saccharomyces Overview
ATUM offers Saccharomyces vectors for cytoplasmic expression with a choice of constitutive (TEF, GPD and ADH) or inducible (GAL1) promoters, and a choice of auxotrophic markers.
Advantages of expression in Saccharomyces cerevisiae:
- Does not require complex media or culture conditions
- Genetically easy to manipulate
- Eukaryotic protein synthesis pathway
- Integrated vectors help genetic stability of the recombinant genes
- Ability to perform higher eukaryotic protein modifications, such as O- or N- linked glycosylation, phosphorylation, disulfide bridge formation, proteolytic processing and folding in a eukaryotic system
- Available as Electra vectors for rapid cloning and testing protein expression in multiple vectors
One undesirable attribute of S. cerevisiae is the potential to hyperglycosylate proteins, which may hinder the antigenic properties or function of the protein by masking key epitopes or functional sites. Additionally, the hyper-antigenic nature of terminal α, 1, 3 glycan linkages added to expressed protein makes them particularly unsuitable for therapeutic use. Finally, S. cerevisiae generally expresses recombinant proteins at relatively lower levels, as compared to Pichia.
Cytoplasmic Protein Expression with Different Promoters
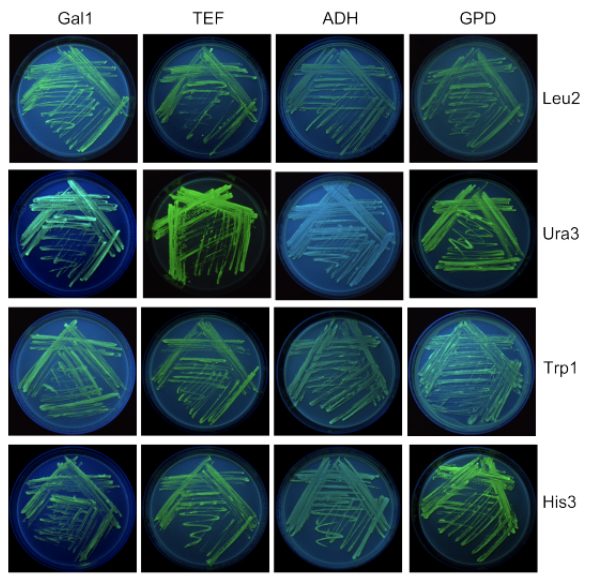
Cytoplasmic DasherGFP expression with various promoters in S. cerevisiae. DasherGFP was cloned into vectors pD1201 (GAL1_P), pD1211 (TEF_P), pD1221 (ADH_P) and pD1231 (GPD_P), all with a Leu2 selectable marker and transformed into S. cerevisiae (MATa/α ura3-52/ura3-52 trp1-289/trp1-289 leu2-3_112/leu2-3_112 his3 Δ1/his3 Δ1). Two clones per construct were grown for 24 hours in complex media, an equivalent amount of culture (approx. 11 OD units at A600) was pelleted. Pellets were resuspended in 250µl Yeast busters reagent. 10µl of lysate was run on SDS-PAGE gel. Quantitation of protein was done by band density compared to BSA as standard. Data is shown as amount of DasherGFP protein expressed per OD unit. Highest constitutive expression was observed with TEF and GPD promoters. ADH is a weak promoter and shows lower expression. Good induction was observed with the GAL1 promoter upon induction with 2% galactose. Expression was almost completely repressed in the presence of glucose.
Cytoplasmic Expression of DasherGFP in S. cerevisiae
Cytoplasmic Expression of DasherGFP in S. cerevisiae. Genes encoding for fluorescent protein (DasherGFP) were cloned in S. cerevisiae vectors (pJ120x-03_Gal1, pJ121x-03_TEF, pJ122x-03_ADH, pJ123x-03_GPD) and transformed into S. cerevisiae (MATa/α ura3-52/ura3-52 trp1-289/trp1-289 leu2-3_112/leu2-3_112 his3 Δ1/his3 Δ1). Plasmid bearing clones were selected by plating on complete CM glucose agar plates with a single amino acid dropout that corresponds to the amino acid selection marker in the plasmid. Plates were incubated for approximately 3 days at 30°C and photographed under UV light. Expression control (untransformed) S. cerevisiae cells are shown on the right.
Secreted Protein Expression
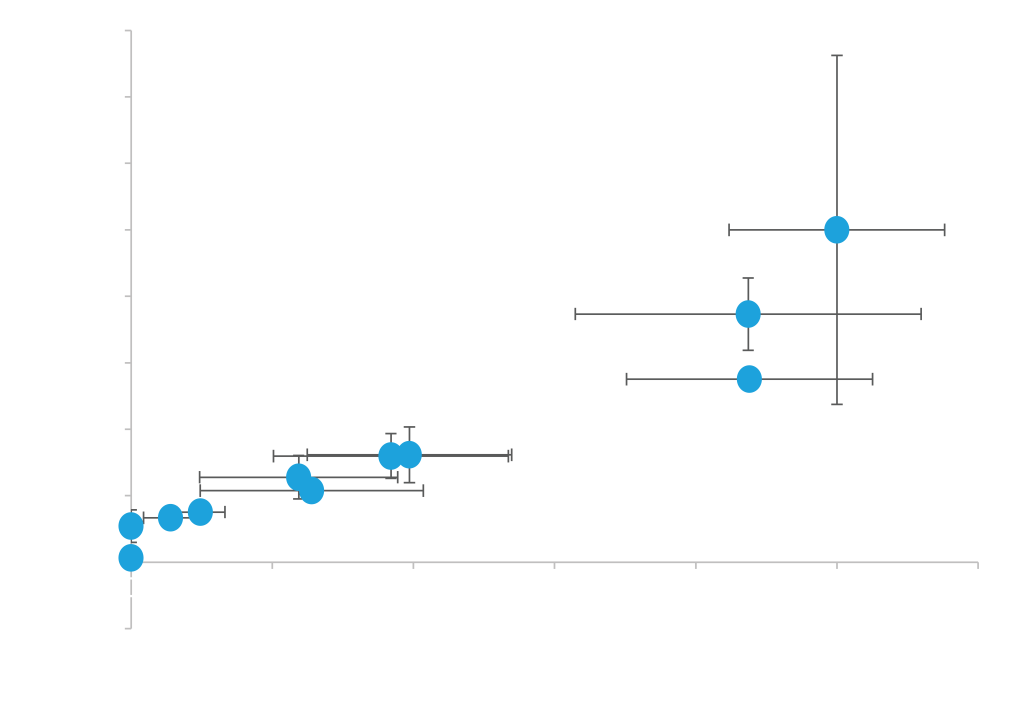
Efficient protein secretion is highly dependent on the combination of the protein and secretion signal used, thus ATUM offers vectors with eleven different secretion signals for targeting proteins to the secretory pathway. You can select vectors from the interactive graphs below, either with the strong constitutive TEF promoter, or the weaker ADH promoter. We recommend testing the entire vector panel to identify the signal that works best with your protein.
Panel A. Different Secretion Signals Work Well with Different Proteins. Secreted Expression of Cutinase and Endopeptidase driven by a constitutive TEF promoter and various secretion signals (pD1214-XX). Expression values shown on x- and y-axis are measurements of expressed protein band densities from a SDS-PAGE gel using BSA as standard. Highest Cutinase expression is observed with either the alpha-factor secretion signal (AKS) or Invertase (IN). Highest Endopeptidase expression is observed with serum albumin (SA) secretion signal. Expression levels range from 6-85 µg/ml for Cutinase and 4-30 µg/ml for Endopeptidase; grown in small batch cultures.
Panel B. Level of Protein Expression can be Modulated by Promoter Strength. Secreted Expression of Cutinase driven by constitutive TEF and ADH promoters (pD1214-XX and pD1221-XX). Expression values shown on x- and y-axis are measurements of expressed protein band densities from a SDS-PAGE gel using BSA as standard. As observed from the graph, relative expression levels of protein with different secretion signals are not influenced by promoter used. TEF is a stronger promoter and shows higher expression levels, while as a weak promoter ADH may be useful for proteins needed at lower expression levels.
References
View Publications using ATUM Saccharomyces Expression Vectors and/or Saccharomyces host systems
Search the ATUM Literature Database, containing over 2,660 scientific publications using ATUM technology for references relevant to your research.
Saccharomyces Protocols
Saccharomyces Culture and Induction Protocol from ATUM
Vector Cost
Notes
- pJ12XX-03C Vectors available as catalog vectors have a DasherGFP® insert, but do NOT contain multiple cloning sites. They can be used as a positive control in conjunction with vectors purchased for gene synthesis orders.
- For cloning of inserts, use Electra pD12XX or pD9XX catalog vectors.
- pD and pM vectors (without an ORF) are provided as linearized DNA in solution (10 reactions). pD or pM vectors with a control ORF are provided as circular plasmids (lyophilized).
- Competent Pichia yeast cell preparation protocol can be found here.
- Synthetic gene sequence must NOT include SacI (AOX1 promoter) or AvrII (GAP1 promoter) as these sites are used during cloning and linearization. If your gene includes these sites it will be cut internally and not cloned correctly.
- Vectors with constitutive promoter result in lower expression levels, but do not require addition of methanol.
The pJ9xx Pichia vectors were constructed by Anton Glieder, et al at the Graz University of Technology in Austria, for protein expression in P. pastoris.